Molecular Docking Analysis | Autodock Results Analysis | Protein Ligand Int | Pymol | LigPlot Etc., - YouTube

AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading - Trott - 2010 - Journal of Computational Chemistry - Wiley Online Library
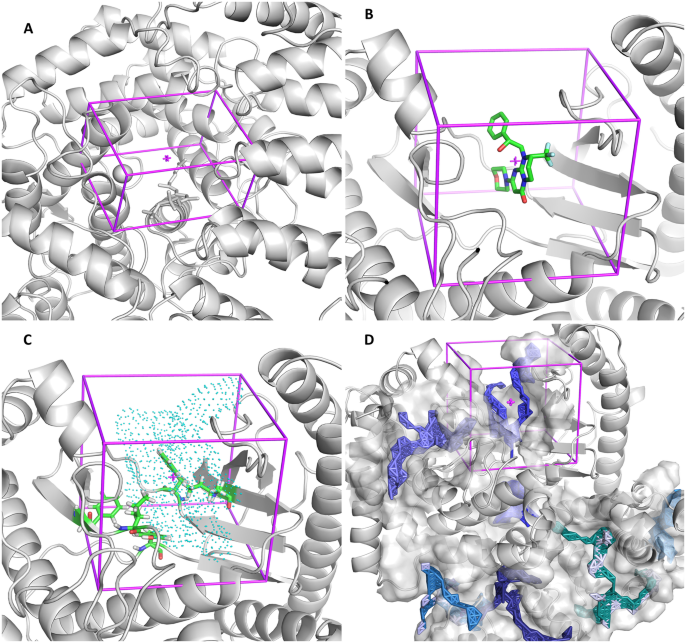
AMDock: a versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4 | Biology Direct | Full Text

Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock - ScienceDirect

CrossDocker: a tool for performing cross-docking using Autodock Vina – topic of research paper in Biological sciences. Download scholarly article PDF and read for free on CyberLeninka open science hub.

Webina: An Open-Source Library and Web App that Runs AutoDock Vina Entirely in the Web Browser | bioRxiv

AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings | Journal of Chemical Information and Modeling